Irepan Salvador-Martínez: Reconstructing and visualising cell lineages
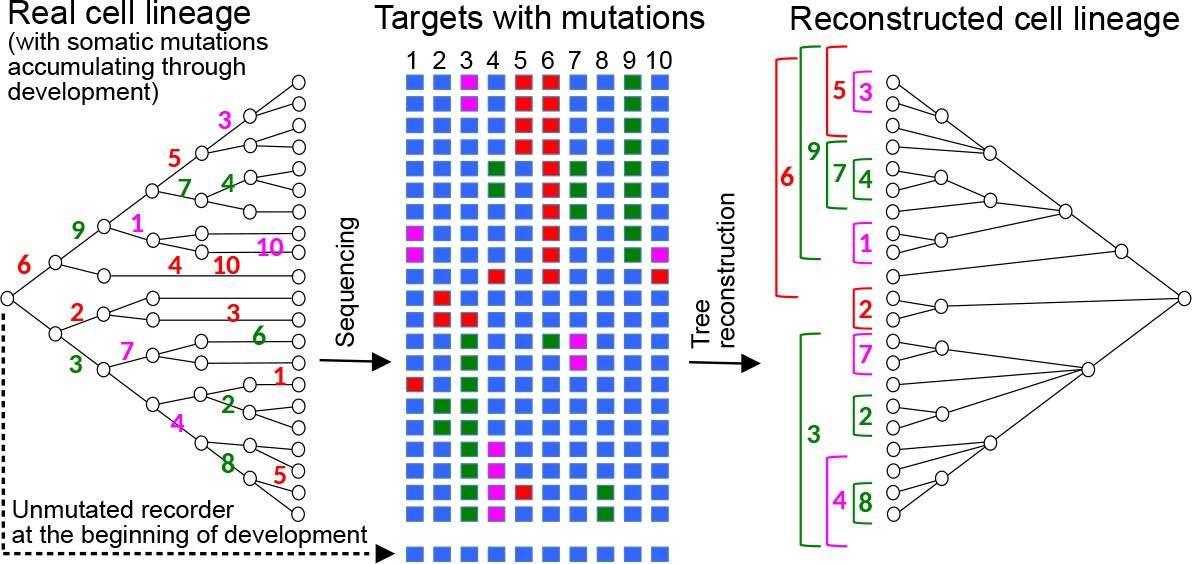
Cell lineages provide the framework for understanding how multicellular organisms are built and how cell fates are decided during development. Describing cell lineages in most organisms is challenging, given the number of cells involved; even a fruit fly larva has approximately 50,000 cells. Recently, the idea of using CRISPR to induce mutations during development as heritable markers for lineage reconstruction has been proposed by several groups. While an attractive idea, its practical value depends on the accuracy of the cell lineages that can be generated by this method. First, I will show results based on computer simulations that incorporate empirical data on CRISPR-induced mutation frequencies in Drosophila. These results show significant impacts from multiple biological and technical parameters – variable cell division rates, skewed mutational outcomes, target dropouts, etc. Our approach reveals the limitations of recently published CRISPR recorders, and indicates how future implementations can be optimised to produce accurate cell lineages. Then, I will present an interactive web-tool for visualising both cell lineage and cell distribution in 3D space. This could be applicable to visualise cell lineages reconstructed by different means, e.g. in situ sequencing of lineage recorders, cell lineage tracking via live imaging, etc. As a proof of concept, I will present an interactive visualisation of different available data, including the cell lineages of the nematode C. elegans and from the limb of a crustacean.
Reference: Salvador-Martínez, I., Grillo, M., Averof, M., & Telford, M. J. (2019). Is it possible to reconstruct an accurate cell lineage using CRISPR recorders? eLife. https://doi.org/10.7554/eLife.40292




